Scientific summary
Proteins do almost all the work that keeps cells – and us – alive. To function properly, proteins need to fold into the correct 3D structure. Achieving the correct fold is difficult in a cell because many alternative outcomes are thermodynamically possible. To this day, a mechanistic understanding of the folding trajectory and maintenance of proteins in live cells remains one of the –if not the– most significant unsolved problem in life science.
To safeguard correct biogenesis of its 20,000 different proteins, (human) cells maintain an intricate network and massive quantity of proteins dedicated to quality control (QC). This QC system allows cells and organisms to cope with varied conditions and to withstand various stresses. The system has limits however, which manifest in disease. In Loss-of-Function (LoF) diseases, such as cystic fibrosis, one specific protein does not function because it fails to adopt its correct structure, resulting in aggregation or degradation. In Gain-of-Toxicity (GoT) diseases, such as Parkinson, proteins assemble to toxic aggregates instead of their functional fold. Due to our limited understanding of the molecular processes involved in protein biogenesis, we lack cures for all but a few protein diseases.
Mission of the program
We take our health for granted until it fails. Human health is completely dependent on protein health in our cells, which is relatively robust because it is managed by an intricate molecular QC network. A limited repertoire of only 10s of different chaperones promotes folding of over 20,000 client proteins with high specificity. The chaperones are complemented by QC factors that catalyze post-translational modifications and degradation. Collectively, these factors form an intricate molecular network that buffers protein homeostasis of humans over dramatically differing conditions over a lifetime.
We notice the molecular QC when specific clients have left the stable trajectories within this complex network. Toxic clumps of misfolded proteins are hallmarks of different neurodegenerative diseases, which affect mostly the elderly. Mutations of specific clients hamper the correct folding of important proteins in congenital diseases of young patients. The QC factors triage the defective protein and determine its fate, be it rescue, degradation, or aggregation. A precise molecular understanding of the QC network is required to tune it for pathological conditions.
Aims of the Program
Achieving the moonshot vision of FLOW for control of the fates of proteins in vivo will require the integrated and dedicated expertise of our multi- and interdisciplinary consortium. The combined expertise ranges from cell biology through biochemistry, molecular biology to biophysics, from theory to experiment, from microscopy to the test tube.
FLOW brings these research experts together in a well-structured program with the joint commitment “to establish how a single protein navigates the cellular quality-control landscape towards health versus disease” and “to use this knowledge to steer the QC-landscape and predetermine the protein’s fate”.
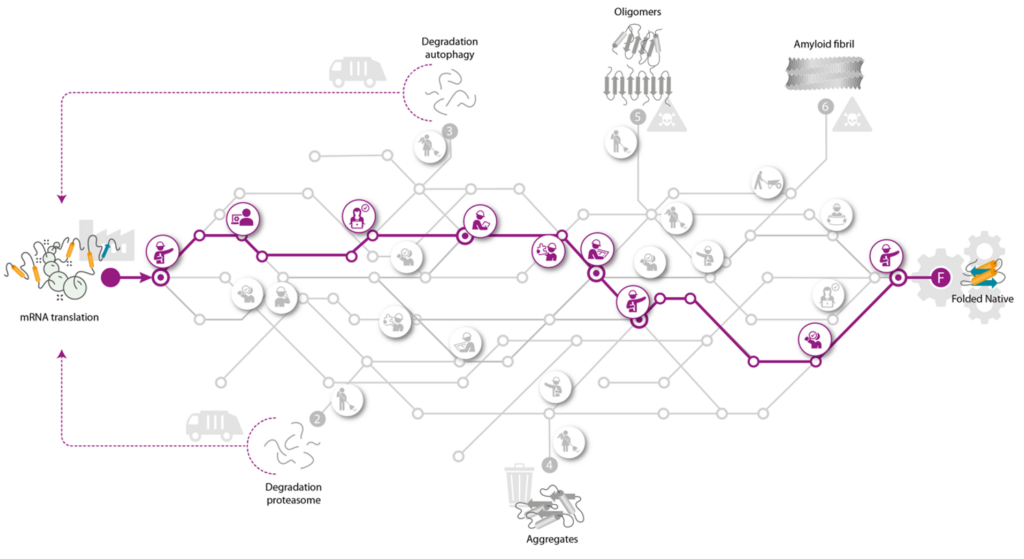
To unravel how misfolded and aggregated proteins can be steered towards fates that are optimal to the health of the cell we will make use of and further develop state of the art technologies that enable us to identify, quantify and characterize the molecular processes that are at the heart of the problem. To map the QC system, with all its QC stations, we selected aSyn and CFTR, as the two model protein clients because they together face every possible fate proteins can experience.
The Overall Aims of the proposal are
- To establish all possible fates for the two model proteins, produce an inventory of their interaction partners with spatio-temporal resolution, and establish the effects of client and QC factor variation on the cell. We will Discover (WP1) which players in the QC system are essential for the client proteins to reach a certain fate. We aim to establish all possible fates for the two model proteins in cells, produce an inventory of their interaction partners with spatio-temporal resolution, and establish the effects of client and QC factor variation on the cell.
- To reconstitute the triage processes for folding versus aggregation or degradation in vitro, to acquire an understanding of decision making towards each fate. We will Rebuild (WP2) the triage processes for folding versus aggregation or degradation in vitro, to acquire an understanding of decision making towards each fate. We use this combined knowledge to develop a predictive model for triage that can be generally applied to protein misfolding related diseases.
- To have the understanding to reach full control over protein fate, in cell, tissue, and organism, and to generalize into a unified theory for protein triage. Our ultimate goal is to aim at obtaining full Control (WP3) over the fate of all proteins and thereby over cell health and as a consequence health at the level of tissues and organisms.
Work Package 1 – DISCOVER
Leading PIs: Braakman, Vertegaal
Associated researchers: Balch, Karras, Taipale, Veltkamp, CFF Labs
Within FLOW we will share all experimental models and systems to unify our data sets, and we will incorporate all data available to us, through publications and collaborations (such as the associated researchers).
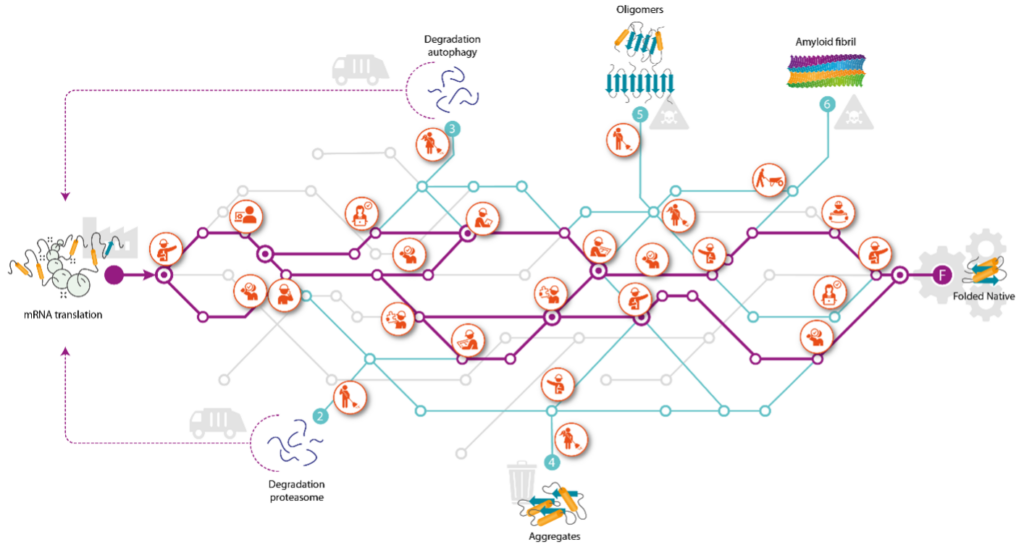
This workpackage aims at understanding the QC network, by identifying the fates and interactions of the two model proteins in cells. FLOW will focus on uncovering the full traffic map for these two at every step of their journey in cells, which starts with synthesis and release from the ribosome, then to intermediate folding, fully native folding, or to misfolding, leading to degradation and/or aggregation. FLOW will chart the cellular QC network with spatiotemporal resolution, including chaperones, co-chaperones, other interaction partners, and post-translational modifications encountered by these proteins during their journeys. Overall goal of FLOW is to exploit this QC network to ultimately obtain full control of the fates of these proteins in cells. FLOW intends to reach such depth of understanding of this journey that it will allow steering of each protein’s fate, leading to fully balanced proteomes required for optimal functioning of cells, preventing proteinopathies.
Work Package 2 – REBUILD (in vitro)
Leading PIs: Claessens, Rüdiger
Overall Aim. To reconstitute the triage processes for folding versus aggregation or degradation in vitro, to acquire an understanding of decision making towards each fate.
This workpackage aims at understanding the QC network at a molecular level, by reconstituting and modulating key steps from purified and well-defined components. FLOW will focus on understanding the decision-making processes within the cellular protein-QC network, which includes the chaperone machinery, ubiquitin-proteasome system, and autophagy machinery. By reconstituting key pathways and controlling the composition of QC machinery and substrates, we will explore the mechanisms that direct proteins towards refolding, disaggregation, or degradation. We will do this at high resolution and at the molecular level. We will examine the roles of post-translational modifications, (co-)chaperones, and client-specific factors in these processes. Goal of FLOW is to build minimal systems that can efficiently manage misfolded or aggregated CTFR and/or α-synuclein, by identifying essential and redundant components, interdependencies, and hierarchical decision-making events. We will also investigate, e.g. by drugs, how the cell influences these decisions and seeks ways to manipulate them for optimal cell and organism health. FLOW ultimately aims to generalize the findings to control the fate of all proteins, thereby improving overall cellular health.
Work Package 3 – CONTROL
Leading PIs: Förster, Hipp
Overall Aim. To have the understanding to reach full control over protein fate, in cell, tissue, and organism, and to generalize into a unified theory for protein triage.