There are currently no open positions available.
Grey title means the position is no longer available
Mapping the proteostasis landscape for CFTR
Principal Investigators: Ineke Braakman and Peter van der Sluijs
Key collaborations: Alfred Vertegaal, Mark Hipp, Arnold Boersma, Friedrich Förster, Harm Kampinga

Main research question
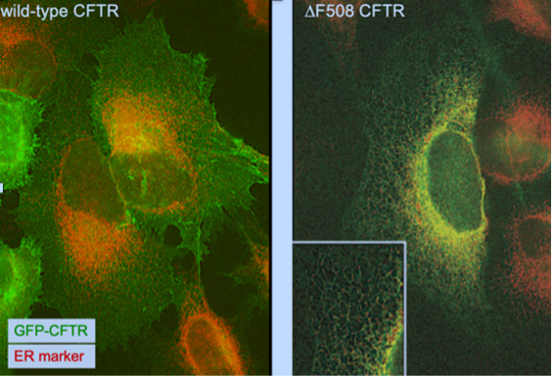
What are the fates of CFTR (the Cystic Fibrosis Transmembrane conductance Regulator) in cell systems and how do these fates change upon altering the proteostasis network?
Background and goals
Accurate and timely delivery of correctly folded proteins is at the heart of cell function. How the folding steps at different locations proceed and are facilitated and coordinated by chaperones is incompletely understood. We will determine the principles underlying folding, misfolding, and disposal of the multispan membrane protein CFTR in cell models. Due to the complex and redundant nature of the quality control (QC) network, different client proteins can be handled by different pathways within the network, resulting in different fates (e.g. degradation by autophagy vs degradation by the ubiquitin proteasome system vs sequestration in protein compartments like the aggresome). In this project, we will characterize the different fates that CFTR can end up in, and determine the pathways that CFTR uses towards these fates. This will help to identify the parts of the QC network that are necessary for a beneficial outcome for wild-type or mutant protein. In these studies we will also include experiments with α-Synuclein in a collaborative project with M. Hipp of FLOW.
Profile/background candidate
We are looking for 2 candidates with a strong background in biochemistry, cell biology, or molecular biology. We welcome post-docs who would like to take the project into independence. Additional positions not directly related to FLOW can be found on our website.
Key references
- Van der Sluijs P, Hoelen H, Schmidt A, Braakman I (2024) The folding pathway of ABC transporter CFTR: effective and robust. J Mol Biol Apr 26: 168591. doi: 10.1016/j.jmb.2024.168591.
- Im J, Hillenaar T, Yeoh HY, Sahasrabudhe P, Mijnders M, van Willigen M, Hagos A, de Mattos E, van der Sluijs P, Braakman I (2023) ABC-transporter CFTR folds with high fidelity through a modular, stepwise pathway. Cell Mol Life Sci. 80: 33. doi: 10.1007/s00018-022-04671-x.
- Kleizen B., van Vlijmen T, de Jonge H, Braakman I (2005). CFTR folds predominantly co-translational. Mol Cell 20:277-87. doi: 10.1016/j.molcel.2005.09.007.
Read more…
Insights into protein aggregate management
Principal Investigator: Mireille Claessens
Key collaborations: Stefan Rüdiger, Evan Spruijt, Anne Wentink, Arnold Boersma

Main research question
How does the protein quality control system target and manage protein aggregates with different biochemical and thermodynamic properties?
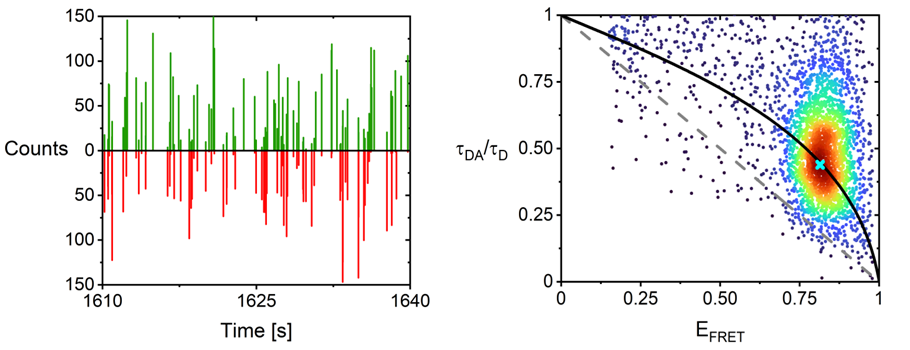
Background and goals
Molecular chaperones have been shown to disaggregate amyloid fibrils. Early protein aggregate species, whether on or off the pathway to forming amyloid fibrils, are thermodynamically less stable and should therefore be easier to disaggregate and refold with molecular chaperones. The goal of this project is to investigate how the protein quality control system targets aggregates of different proteins and different aggregate species in reconstituted model systems. How aggregation, disaggregation, and refolding are influenced by physicochemical changes, such as post-translational modifications or other alterations in the physicochemical environment will also be explored. Furthermore, in this project we will identify the system’s limitations, determine the causes of these limits, and explore their implications. The findings will be related to, and verified in cells. The ultimate goal is to use this combined information to understand how the fate of protein aggregates is controlled.
Molecular chaperones have been shown to disaggregate amyloid fibrils. Early protein aggregate species, whether on or off the pathway to forming amyloid fibrils, are thermodynamically less stable and should therefore be easier to disaggregate and refold with molecular chaperones. The goal of this project is to investigate how the protein quality control system targets aggregates of different proteins and different aggregate species in reconstituted model systems. How aggregation, disaggregation, and refolding are influenced by physicochemical changes, such as post-translational modifications or other alterations in the physicochemical environment will also be explored. Furthermore, in this project we will identify the system’s limitations, determine the causes of these limits, and explore their implications. The findings will be related to, and verified in cells. The ultimate goal is to use this combined information to understand how the fate of protein aggregates is controlled.
Key techniques
This project strongly relies on advanced (single molecule) fluorescence microscopy and spectroscopy approaches such as sm-FRET, TCCD, sm-burst analysis, FCS. Additionally biophysical methods and biochemical approaches will be used.
Profile/background candidate
We are looking for candidates with a background in either biotechnology/biochemistry with experience with (some of) the above mentioned microspectroscopy methods or a background in (bio)physics/physical chemistry with experience in studying biomacromolecules.
Key references
- Heesink, G.; van den Oetelaar, M. C. M.; Semerdzhiev, S. A.; Ottmann, C.; Brunsveld, L.; Blum, C.; Claessens, M. M. A. E., 14-3-3τ as a Modulator of Early α-Synuclein Multimerization and Amyloid Formation. ACS Chemical Neuroscience 2024.
- Heesink, G.; Marseille, M. J.; Fakhree, M. A. A.; Driver, M. D.; van Leijenhorst-Groener, K. A.; Onck, P. R.; Blum, C.; Claessens, M., Exploring Intra- and Inter-Regional Interactions in the IDP α-Synuclein Using smFRET and MD Simulations. Biomacromolecules 2023, 24 (8), 3680-3688.
- Fakhree, M. A. A.; Nolten, I. S.; Blum, C.; Claessens, M., Different Conformational Subensembles of the Intrinsically Disordered Protein α-Synuclein in Cells. Journal of Physical Chemistry Letters 2018, 9 (6), 1249-1253.
- Fakhree, M. A. A.; Konings, I. B. M.; Kole, J.; Cambi, A.; Blum, C.; Claessens, M., The Localization of Alpha-synuclein in the Endocytic Pathway. Neuroscience 2021, 457, 186-195.
Read more…
Imaging CFTR and a-synuclein biogenesis in situ
Principal Investigator: Friedrich Förster
Key collaborations: Ineke Braakman, Alfred Vertegaal, Peter van der Sluijs, Mark Hipp, Martijn Huynen

Main research question
Which interactors trigger distinct fates of CTFR and a-synuclein in the cell and how?
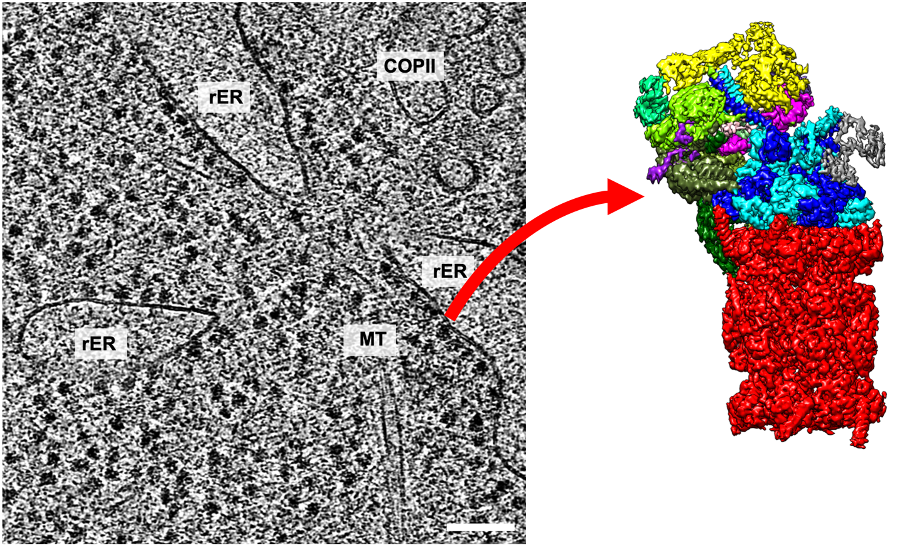
Background and goals
High resolution imaging in native or close-to-native environments reveals the molecular details of interactions that govern the fate of proteins. To this end, cryo-electron tomography plays a unique role due to its unique resolving power. The Förster lab pioneered the use of cryo-ET to investigate distinct states of the protein biogenesis machinery at the Endoplasmic reticulum. The aim is now to expand on these studies to explore the distinct fates of CTFR and a-synuclein in the cellular context. Cryo-ET studies will be combined with cellular engineering, fluorescence light microscopy, mass-spectrometry characterization and protein complex modelling. Primary aims will be to capture premature degradation of CFTR and its molecular facilitators in the acts, as well as revealing key aspects of the a-syn aggregation pathway in situ.
Key techniques
Cryo-electron tomography, protein assembly structure modelling, cell culture, fluorescence light microscopy, genetic engineering, protein chemistry.
Profile/background candidate
We are looking for candidates with a background in biochemistry, biophysics or biology.
Key references
- Fedry J, Silva J, Vanevic M, Fronik S, Mechulam Y, Schmitt E, des Georges A, Faller WJ, Förster F, Visualization of translation reorganization upon persistent ribosome collision stress in mammalian cells. Mol. Cell 84:1078-1089 e4 (2024). https://doi.org/10.1016/j.molcel.2024.01.015
- Gemmer M, Chaillet ML, van Loenhout J, Cuevas Arenas R, Vismpas D, Grollers-Mulderij M, Koh FA, Albanese P, Scheltema RA, Howes SC, Kotecha A, Fedry J, Förster F, Visualization of translation and protein biogenesis at the ER membrane. Nature 614:160-167 (2023). https://doi.org/10.1038/s41586-022-05638-5
- Liaci AM, Steigenberger B, Telles de Souza PC, Tamara S, Grollers-Mulderij M, Ogrissek P, Marrink SJ, Scheltema RA, Förster F, Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol. Cell 81:3934-3948 (2021). https://dspace.library.uu.nl/bitstream/handle/1874/412779/1_s2.0_S1097276521006006_main.pdf?sequence=1&isAllowed=y
Read more…
Mapping the proteostasis landscape for alpha-synuclein
Principal Investigator: Mark Hipp
Key collaborations: Alfred Vertegaal, Arnold Boersma, Ineke Braakman, Friedrich Foerster, Harm Kampinga, Peter van der Sluijs

Main research question
Define the potential fates of mutant and wildtype alpha-synuclein in cellular systems, and change it by altering the proteostasis network.
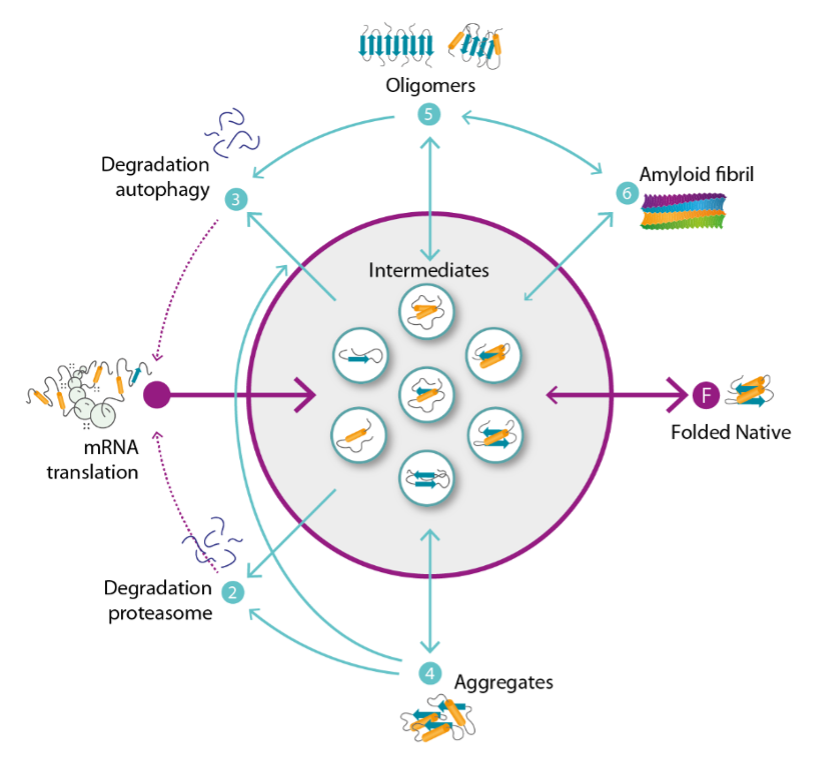
Background and goals
Due to the complex and redundant nature of the quality control (QC) network, many client proteins can be handled by different pathways within the network, resulting in different fates (e.g. degradation by autophagy vs degradation by the ubiquitin proteasome system vs sequestration in protein compartments like the aggresome). In this project we characterize the different fates that alpha-synuclein can end up in in cellular systems, and determine the pathways that it uses to navigate across the QC landscape towards these fates. This will help us to identify the parts of the QC network that are necessary for a beneficial outcome for both wildtype or mutant protein.
Profile/background candidate
We are looking for candidates with a background in cell biology and cellular biochemistry, that are able to closely collaborate with the groups specialized in mass-spectrometry and electron microscopy.
Key references
- Trinkaus VA, Riera-Tur I, Martínez-Sánchez A, Bäuerlein FJB, Guo Q, Arzberger T, Baumeister W, Dudanova I, Hipp MS, Hartl FU, Fernández-Busnadiego R. In situ architecture of neuronal α-Synuclein inclusions. Nat Commun. 2021;12(1):2110. doi: 10.1038/s41467-021-22108-0.
- Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019; 365(6451):342-347.
- Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, Hipp MS Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016; 351(6269):173-6.
Read more…
Decision-making on the fate of damaged proteins
Principal Investigator: Stefan Rüdiger
Key collaborations: Mireille Claessens, Kasia Tych, Anne Wentink (project 1);
Mireille Claessens, Alfred Vertegaal, Monique Mulder, Friedrich Förster (project 2)

Main research questions
1. How do molecular chaperones control the fate of damaged proteins?
2. How do shape and properties of protein aggregates determine their fate?
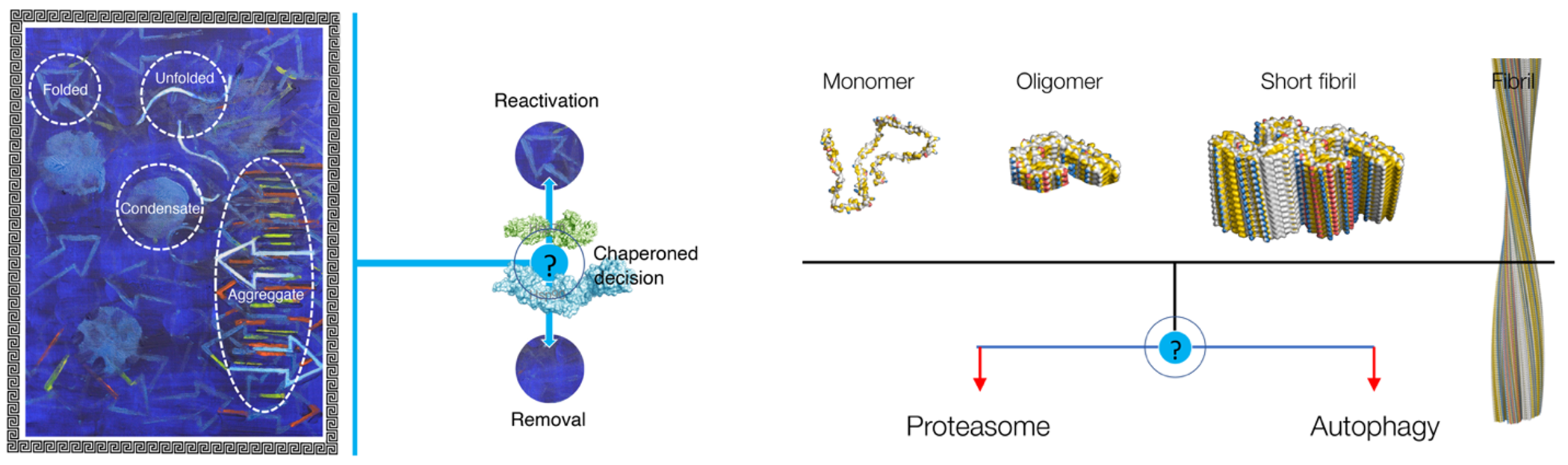
Background and goals (project 1)
The aim of this project is to uncover how the central decision-making node between (re)folding and degradation works. The Hsp70-Hsp90 axis constitute the central, universally conserved a chaperone axis. Key components of this node are the conserved Hsp70 and Hsp90 chaperone systems. This axis shuttles misfolded proteins between (re)folding, reactivation and degradation. The Rüdiger group established that Hsp90 acts downstream of Hsp70, releasing an Hsp70-inflected folding block. This offers the opportunity for a plethora of co-chaperones to tweak the system. The molecular basis for the modulation of the system is poorly understood. This project aims to uncover the molecular mechanism of decision-making between reactivation and removal. We use as paradigmatic substrates a-synuclein fibrils and soluble, aggregation-prone CFTR domains. We aim to deliver a long-standing dream of the molecular life sciences, which is to understand the system to such an extent that we can rebuild the key component and to modulate the chaperone machinery at will.
Background and goals (project 2)
This project aims to determine the relation of the shape of damaged protein and the degradation channel. The Rüdiger group established the use of FIDA technology in combination with fibril paint to monitor the size of damaged protein. This now allows for the first time a quantitative, substrate driven view to determine their fate. The project will establish the influence of size, length and shape of protein aggregates in the triaging of degradation. The goal is to identify the entry criteria for degradation by the proteasome and by autophagy, and how cellular factors and drugs can influence this decision. The project centres chiefly on a-synuclein fibrils, while misfolded CFTR domains serve as comparison.
Key techniques
Protein chemistry and biophysical methods, including microscale methods such as FIDA, thermophoresis, sensitive fluorescence methods, EM and light scattering techniques.
Profile/background candidate
We are seeking candidates with background in (bio)chemistry, (bio)physics, medical biology or related fields.
Key references
- Aragonès Pedrola J, Dekker FA, Garfagnini T, Mayer G, Koopman MB, Bergmeijer M, Förster F, Hoozemans JJM, Jensen H, Friedler A#, Rüdiger SGD#. Fibril Paint to detect Amyloids and determine Fibril Length. bioRxiv 2023; 10.13.562220. doi: https://doi.org/10.1101/2023.10.13.562220
- Koopman MB, Ferrari L, Rüdiger SGD#. How do protein aggregates escape quality control in neurodegeneration? Trends Neurosci. 2022, 45, 257-271. doi: 10.1016/j.tins.2022.01.006
- Ferrari L, Stucchi R, Konstantoulea K, van de Kamp G, Kos R, Geerts WJC, Bezouwen LS, Förster FG, Altelaar M, Hoogenraad CC, Rüdiger SGD#. Arginine pi-stacking drives binding to fibrils of the Alzheimer protein Tau. Nature Comm. 2020; 11:571. doi: 10.1038/s41467-019-13745-7
- Morán Luengo T, Mayer MP, Rüdiger SGD#. The Hsp70-Hsp90 Chaperone Cascade in Protein Folding. Trends Cell Biol. 2019. 29:164-177.
doi: 10.1016/j.tcb.2018.10.004. Epub 2018 Nov 28. PMID: 30502916. - Morán Luengo T, Kityk R, Mayer MP#, Rüdiger SGD#. Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol Cell. 2018;70:545-552. doi: 10.1016/j.molcel.2018.03.028
Read more…
Ubiquitin-proteasome mediated degradation of misfolded proteins
Principal Investigator: Alfred Vertegaal
Key collaborations: Ineke Braakman, Mark Hipp, Friedrich Foerster, Monique Mulder, Martijn Huynen, Peter van der Sluijs

Main research questions
How does the ubiquitin-proteasome system control the fate of damaged proteins to prevent protein aggregation?
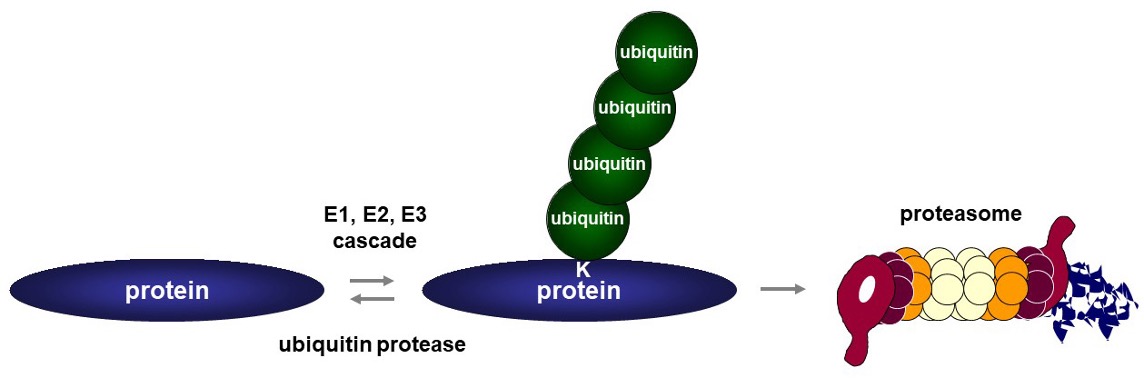
Background and goals
Irreversibly misfolded proteins are ubiquitinated and subsequently degraded by the proteasome. The Vertegaal group has ample experience with ubiquitin and ubiquitin-like protein modification and has uncovered extensive sets of target proteins for the ubiquitin-proteasome system. The molecular basis for the decision to refold or degrade misfolded proteins remains poorly understood. This project aims to uncover the molecular mechanism of decision-making on proteasomal degradation versus refolding of misfolded proteins. We use as paradigmatic substrates a-synuclein fibrils and CFTR. Our main aim is to harness the ubiquitin-proteasome system to fully control protein degradation.
Key techniques
Functional molecular cell biology, protein chemistry, proteomics, mass spectrometry, bioinformatics.
Profile/background candidate
We are looking for candidates with a background in functional molecular cell biology, cellular biochemistry and and/or mass-spectrometry who are able to work closely with the collaborating groups in the network.
Key references
- Trulsson F, Akimov V, Robu M, van Overbeek N, Berrocal DAP, Shah RG, Cox J, Shah GM, Blagoev B, Vertegaal ACO. Deubiquitinating enzymes and the proteasome regulate preferential sets of ubiquitin substrates. Nat Commun. 2022 May 18;13(1):2736. doi: 10.1038/s41467-022-30376-7.
- Liebelt F, Sebastian RM, Moore CL, Mulder MPC, Ovaa H, Shoulders MD, Vertegaal ACO. SUMOylation and the HSF1-Regulated Chaperone Network Converge to Promote Proteostasis in Response to Heat Shock. Cell Rep. 2019 Jan 2;26(1):236-249.e4. doi: 10.1016/j.celrep.2018.12.027.
- Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, Vertegaal ACO. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics. 2008 Nov;7(11):2107-22. doi: 10.1074/mcp.M800025-MCP200.
Read more…
Sensors to investigate chaperone-client interactions in cells and in vitro compartments
Principal Investigator: Arnold Boersma
Key collaborations: Anne Wentink, Alfred Vertegaal, Mark Hipp

Main research question
Can we design sensors that measure which chaperones interact when and where with their clients in cells? Can we also use these for high-throughput screening in vitro?
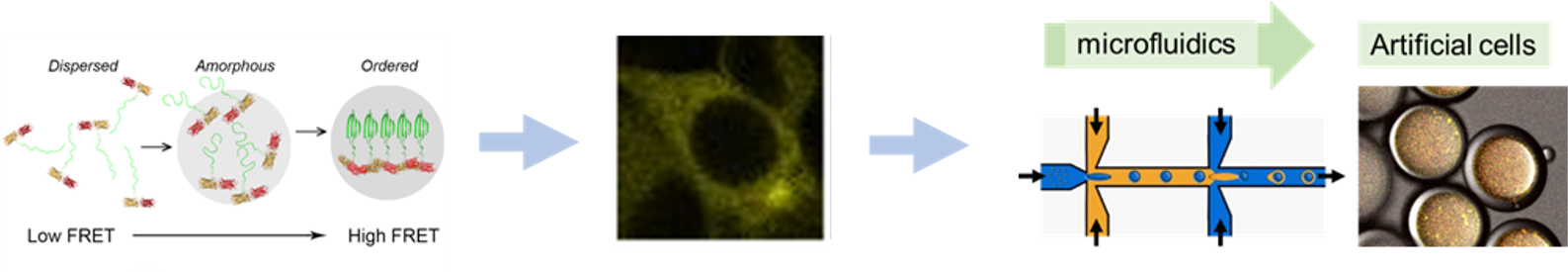
Background and goals
It is unclear why the cell uses so many different chaperones and who interacts when and where with a particular client protein. You aim to solve this riddle by 1) creative design of novel genetically encoded sensors to quantify interactions between chaperones and clients and 2) adapting a microfluidic platform that allows you to reconstruct many different chaperone-client scenarios in artificial cells.
Fluorescence-based genetically encoded sensors allow sensitive measurement with minimal perturbations and exceptionally high spatial and temporal resolution. You will use your creativity, intuition, and software to engineer the sensors that allow you to monitor new interactions in cells. Potential interactions are informed by mass spectrometry measurements. By placing these interactions of purified proteins in droplet microfluidics, which provides precise control over compartmentalized volume and content in a high-throughput and reproducible manner, you arrive at a better understanding of the client’s interactions.
Key techniques
- Microfluidics
- Fluorescence (high-throughput and confocal) measurement techniques
- Protein expression, handling, and (biophysical and biochemical) analysis
Profile/background candidate
You have completed a master’s or a Ph.D. in biochemistry, molecular biology, biophysics, analytical chemistry, or a related research field. You enjoy making new proteins that no one has made before and applying these to important biological scenarios.
Key references
On our microfluidics:
- High macromolecular crowding in liposomes from microfluidics. LPB Guerzoni, AVC de Goes, M Kalacheva, J Hadula, M Mork, L De Laporte, AJ Boersma. Advanced Science 2022: 9 (27), 2201169
Examples of our probes:
- A FRET-based method for monitoring structural transitions in protein self-organization. Q Wan, SN Mouton, LM Veenhoff, AJ Boersma. Cell Reports Methods 2022: 2 (3), 100184
- A sensor for quantification of macromolecular crowding in living cells. Boersma AJ, Zuhorn IS, Poolman B. Nat Methods. 2015: 227-9
Read more…
Explaining the origin of chaperone system complexity using systems biology
Principal Investigator: Martijn Huynen
Key collaborations: Evan Spruit, Harm Kampinga, Mark Hipp, Alfred Vertegaal

Main research question
Why are there, in human, so many different copies of the chaperone system proteins, and what determines their specificity for their clients?
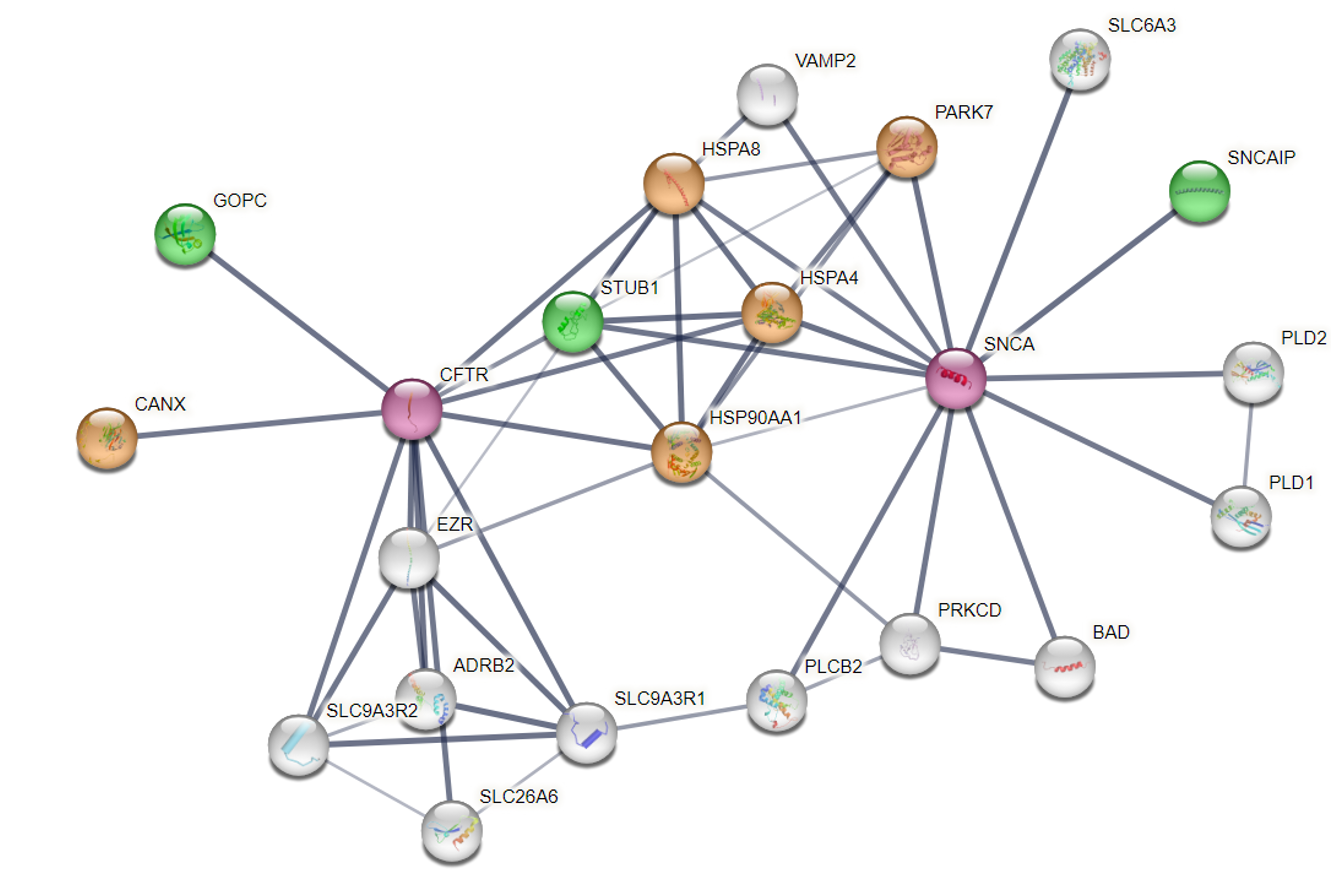
Background and goals
With at least 14 HSP70 genes, 36 HSP40 genes, 12 NEFs, 5 HSP90 genes and many co-factors like 10 small HSPBs and several chaperonines (GroEL/ES, TRiC), the human genome harbours a very versatile set of chaperones. Although some of the plenitude of these chaperones can be attributed to their functioning in different compartments, most of it is not understood. The goal of this project is to derive specific, testable hypotheses to explain the apparent redundancy of the human chaperone system. What are the preferred interaction combinations of the chaperones with their co-chaperones and their clients, and how did they evolve? For this we will use computational methods to exploit the accumulation of sequenced genomes, (predicted) 3D structures, (single cell) co-expression data and interaction data as well as proteomics results from crispr-cas screens executed in the consortium. We will delineate functionally relevant evolutionary information like the evolutionary origin and loss of the chaperones and their clients, and the (co)evolution of the interaction interfaces of chaperone proteins. We validate most promising predictions by targeted interaction measurements, like measuring the solubility or functionality of client proteins after knockout of their chaperone, fluorescence microscopy and proximity labelling, which we will perform within the consortium.
The work builds upon the in-depth evolutionary analyses of Martijn Huynen who has elucidated the (co) evolution of molecular systems in mitochondria and cilia and predicted new members of such systems using co-evolution and co-expression, and who has recently shown how the appearance of a new gene can compensate for the evolutionary degeneration of another one.
Key techniques
Bioinformatics, sequence analysis, including co-evolution and its applications e.g. in AlphaFold-Multimer, computational analysis of gene expression data and proteomics data.
Profile/background candidate
We are looking for a candidate with experience in analysis of molecular evolution, gene expression and other omics data, and affinity with large scale computational analyses.
Key references
- Joeri van Strien, Felix Evers, Madhurya Lutikurti, Stijn L Berendsen, Alejandro Garanto, Geert-Jan van Gemert, Alfredo Cabrera-Orefice, Richard J Rodenburg, Ulrich Brandt, Taco W A Kooij, Martijn A Huynen Plos Comp. Biol. 2023 19(8):e1011090 Comparative Clustering (CompaCt) of eukaryote complexomes identifies novel interactions and sheds light on protein complex evolution.
- Vivek Singh, J. Conor Moran, Yuzuru Itoh, Iliana C. Soto, Flavia Fontanesi, Mary Couvillion, Martijn A. Huynen, Stirling Churchman, Antoni Barrientos, Alexey Amunts bioRxiv 2022.06.20.496763. Structural basis of LRPPRC-SLIRP-dependent translation by the mitoribosome.
- Teunis JP van Dam TJ, Matthew J Townsend, Martin Turk, Avner Schlessinger, Andrej Sali, Mark C Field, Martijn A Huynen. Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):6943-8. Evolution of modular intraflagellar transport from a coatomer-like progenitor.
Read more…
Balancing protein degradation to prevent toxic aggregate accumulation
Principal Investigator: Monique Mulder
Key collaborations: Alfred Vertegaal, Stefan Rüdiger, Mark Hipp

Main research question
How does the ubiquitin-proteasome system orchestrate the fate of damaged proteins to avert protein aggregation?
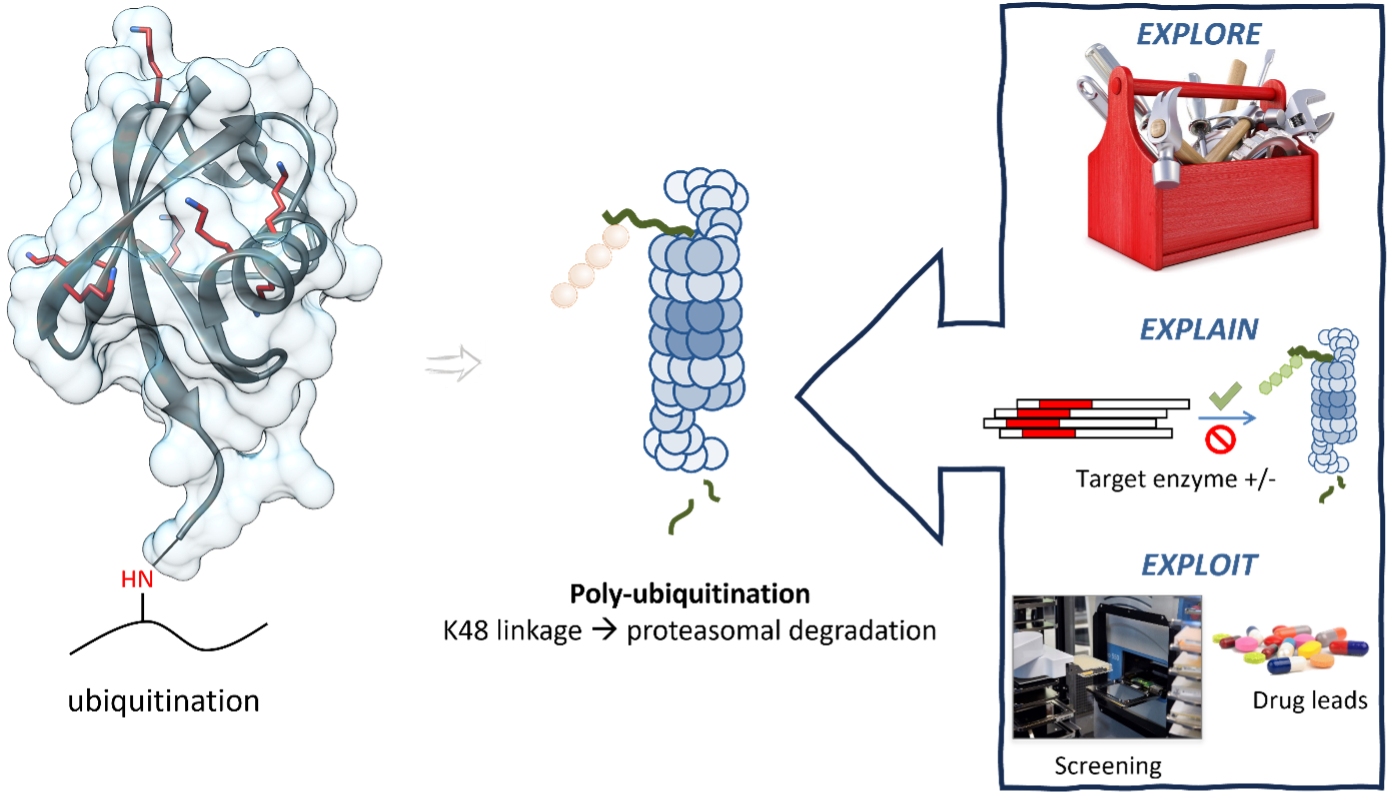
Background and goals
To address this question, we propose to investigate the roles of specific members of the ubiquitin-modulating enzyme families, including the ‘writers’ and ‘erasers,’ in regulating ubiquitination events. By interfering with these targeted ubiquitination processes, we aim to elucidate the mechanisms by which the ubiquitin-proteasome system safeguards against protein aggregation and maintains proteostasis.
To achieve this, we employ cutting-edge synthetic tools such as ubiquitin-based chemical probes. These tools enable us to visualize transient ubiquitylation intermediates, facilitating the identification of potential therapeutic targets. By elucidating the precise points at which these intermediates influence protein fate—whether towards aggregation or degradation—we aim to provide insights into the underlying molecular pathways. Furthermore, by investigating enzymatic cascades and developing small molecule modulators, we endeavor to uncover molecular pathogenesis and pioneer innovative therapeutic strategies.
Key techniques
Peptide and Protein Chemistry, Chemical biology, High-throughput screening, Organic and medicinal chemistry.
Profile/background candidate
We are looking for a candidate with a background in chemistry and/or chemical biology.
Key references
- Pérez Berrocal DA, Vishwanatha TM, Horn-Ghetko D, Botsch JJ, Hehl LA, Kostrhon S, Misra M, Ðikić I, Geurink PP, van Dam H, Schulman BA, Mulder MPC. A Pro-Fluorescent Ubiquitin-Based Probe to Monitor Cysteine-Based E3 Ligase Activity. Angew Chem Int Ed Engl. 2023 Aug 7;62(32):e202303319. doi: 10.1002/anie.202303319.
- Mons E, Kim RQ, van Doodewaerd BR, van Veelen PA, Mulder MPC, Ovaa H. Exploring the Versatility of the Covalent Thiol-Alkyne Reaction with Substituted Propargyl Warheads: A Deciding Role for the Cysteine Protease. J Am Chem Soc. 2021 May 5;143(17):6423-6433. doi: 10.1021/jacs.0c10513.
- Witting KF, van der Heden van Noort GJ, Kofoed C, Talavera Ormeño C, El Atmioui D, Mulder MPC, Ovaa H. Generation of the UFM1 Toolkit for Profiling UFM1-Specific Proteases and Ligases. Angew Chem Int Ed Engl. 2018 Oct 22;57(43):14164-14168. doi: 10.1002/anie.201809232.
Read more…
Protein aggregation and chaperone activity in biomolecular condensates
Principal Investigator: Evan Spruijt
Key collaborations: Mireille Claessens, Stefan Rüdiger, Arnold Boersma, Sonja Schmid

Main research question
How does the local environment created by LLPS affect the triage of damaged or aggregation-prone proteins, and the downstream pathways?
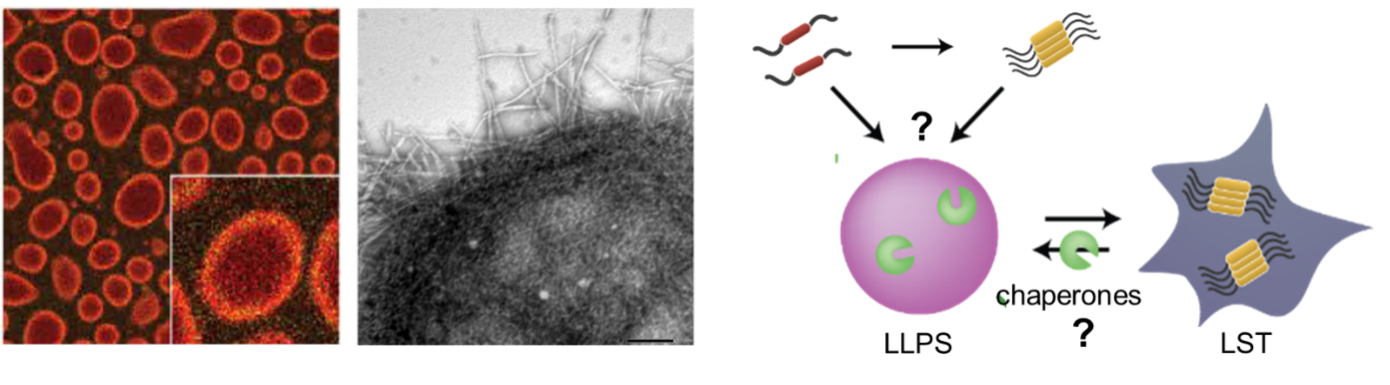
Background and goals
Biomolecular condensates formed by liquid-liquid phase separation (LLPS) are key organizers of biomolecular processes in healthy cells. There is increasing evidence that condensates are also linked to neurodegenerative diseases involving protein misfolding and aggregation, such as Alzheimer’s and Parkinson’s disease, but the underlying mechanisms remain elusive. The Spruijt group established a platform to investigate protein aggregation and chaperone activity in the presence of designer condensates, and showed that condensates can both accelerate and suppress aggregation of paradigmatic protein α-synuclein. This project aims to uncover how chaperones, including Hsp70 and Hsp90, act in and on condensates to triage, prevent or revert protein misfolding and aggregation. We aim to deliver a long-standing dream of life sciences to modulate the chaperone machinery on demand by redesigning, rebuilding and repurposing key components.
Key techniques
Protein chemistry, biophysics, in vitro reconstitution of biomolecular condensates, quantitative fluorescence microscopy, FRET, FLIM and FRAP, light scattering.
Profile/background candidate
We are looking for a candidate with a background in biochemistry, biophysics, chemical biology or a related discipline.
Key references
- Lipiński WP, Visser BS, Robu I, Fakhree MAA, Lindhoud S, Claessens MMAE, Spruijt E#. Biomolecular condensates can both accelerate and suppress aggregation of α-synuclein. Science Advances 2022; 8, abq6495.
- Visser BS, Lipiński WP, Spruijt E#. The role of biomolecular condensates in protein aggregation. Nature Rev. Chem. 2024; accepted.
- Van Haren MHI, Visser BS, Spruijt E#. Probing the surface charge of condensates using microelectrophoresis. Nature Comm. 2024; 15, 3564. DOI: 10.1038/s41467-024-47885-2.
Read more…
Unravelling chaperone function on the single-molecule level
Principal Investigator: Kasia Tych
Key collaborations: Mireille Claessens, Stefan Rüdiger, Anne Wentink

Main research question
How do chaperones control the fate of damaged proteins?
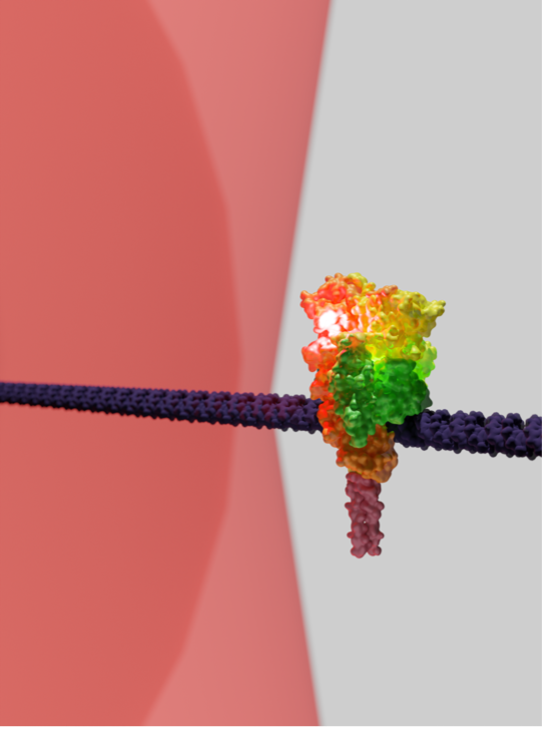
Background and goals
The molecular chaperone and heat-shock protein Hsp90 plays a crucial role in folding and maturing around 20% of all proteins within our cells. This makes it a central component of cellular metabolism, impacting human health significantly by contributing to various cancers, neurodegenerative conditions, and more. Despite its essential function, many fundamental questions about Hsp90’s mechanisms remain unanswered, including an understanding and quantification of the specific tasks Hsp90 carries out to chaperone its clients, alongside its cochaperones, at the molecular level. The KT lab has established methods to address these questions using the optical tweezers and mass photometry.
The overall goal of the project is to use in vitro single-molecule measurements to reconstitute and characterise the Hsp90 chaperone and its interacting cochaperones to obtain mechanistic insights into how they coordinate and act on the two client proteins that are the focus of the FLOW consortium, alpha-synuclein and the cystic fibrosis transport protein, CFTR.
Key techniques
Optical tweezers, mass photometry, protein expression, purification and functionalisation, standard biochemical assays to assess function.
Profile/background candidate
We are looking for a candidate with a background in biochemistry, biophysics or medical biology.
Key references
- Tych, K.M.,Jahn, M, Gegenfurtner, F., Hechtl, V.K., Buchner, J. and Rief, M., Nucleotide-Dependent Dimer Association and Dissociation of the Chaperone HSP90. J Phys Chem B, 2018. 122:11373-80.
- Mondol, T., Silbermann, L.-M., Schimpf, J., Vollmar, L., Hermann, B., Tych, K.M. and Hugel, T., Aha1 regulates Hsp90’s conformation and function in a stoichiometry-dependent way, Biophys. J, 2023. 122: 3458-3468
- Jahn, M. and Tych, K.M., Girstmair, H., Steinmaßl, M., Hugel, T., Buchner, J. and Rief, M., Folding and domain interactions of three orthologs of Hsp90 studied by single-molecule force spectroscopy, Structure, 2018, 26:96-105
Read more…
Chaperones: cooperation to tackle aggregation
Principal Investigator: Anne Wentink
Key collaborations: Arnold Boersma, Mireille Claessens, Stefan Rüdiger

Main research question
How do chaperones prevent and reverse pathological protein aggregation?
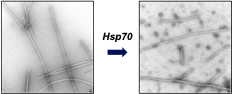
Background and goals
Protein aggregation is a key driver of human disease including neurodegeneration. In response, cells have evolved elaborate networks of molecular chaperones that combat protein aggregation by stabilising and refolding misfolded proteins and preventing aggregation. More surprisingly, the Hsp70 chaperone can also actively disassemble existing protein aggregates. The question remains how Hsp70 toggles between these different cellular functions?
This project aims to understand on a molecular level how different (co-)chaperones enable Hsp70 to perform distinct functions in the folding and aggregation of the model proteins a-synuclein and CFTR and explore how we may selectively tune these functions to combat protein aggregation in disease.
Additional positions are available on related projects. Please contact Anne Wentink for more information.
Key techniques
Protein chemistry and biophysical methods including solution state NMR spectroscopy, fluorescence spectroscopy and electron microscopy.
Profile/background candidate
We are looking for a candidate with a master’s degree in biochemistry, biophysics or a related subject.
Key references
- Wentink A, and Rosenzweig R, Protein disaggregation machineries in the human cytosol, Curr Opin Struct Biol. 2023 Dec; 83 (102735).
- Wentink AS, Nillegoda NB, Feufel J, Ubartaite G, Schneider CP, De los Rios P, Hennig J, Barducci A, Bukau B, Molecular dissection of amyloid disaggregation by the human Hsp70 chaperone, Nature. 2020 Nov;587(7834):483-488.
- Faust O, Abayev M, Wentink A, Maurer M, Nillegoda NB, Bukau B, Rosenzweig R, Hsp40s employ class-specific regulation to drive Hsp70 functional diversity, Nature. 2020 Nov;587(7834) :489-494.
Read more…